Abstract
INTRODUCTION: Myelodysplastic syndromes (MDS) and chronic myelomonocytic leukemia (CMML) are heterogeneous myeloid neoplastic disorders characterized by ineffective hematopoiesis leading to cytopenias and increased risk of transformation to acute myeloid leukemia (AML) . The hypomethylating agents (HMA) azacitidine (AZA) and decitabine (DAC) improve the natural history of MDS and CMML patients (Fenaux et al 2009). However, over half of the cases experience primary failure defined by a lack of response to HMA treatment which is associated with poor prognosis and a median survival of 4-6 months (Garcia-Manero et al. 2016, Jabbour et al. 2010). The highly heterogeneous pathophysiology of myeloid neoplasms and poorly understood mechanisms underlying therapeutic action of HMAs pose substantial challenges in understanding the biology of HMA failure in MDS and CMML.
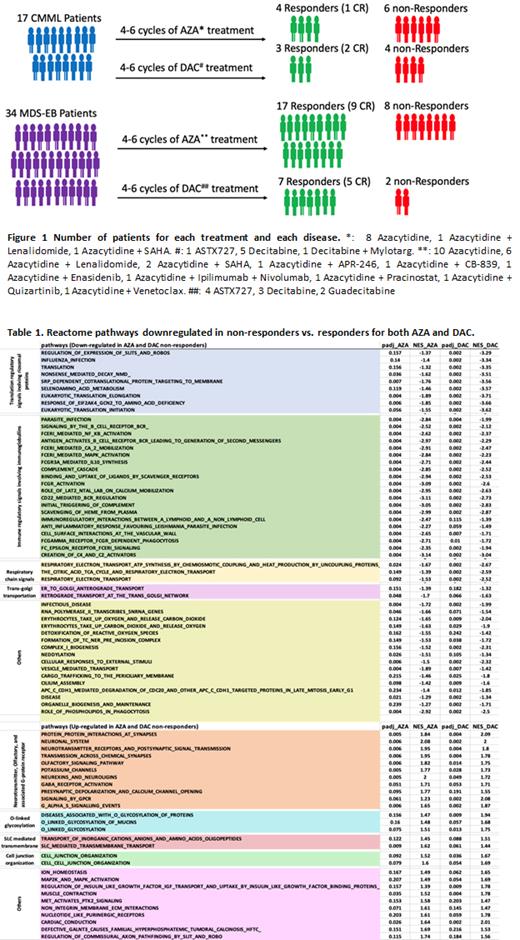
METHODS: We established a cohort of baseline bone marrow (BM) cells that were collected from 17 CMML and 34 MDS patients with excessive blasts (MDS-EB) prior to their HMA based therapies (Figure 1). RNA-Seq based transcriptomic analysis was performed in CD34+ BM hematopoietic stem and progenitor cells (HSPCs) of patients, known to be the cellular origin of these diseases, to identify biological signatures of primary HMA resistance.
RESULTS: Similar to the recent reports regarding the lack of common gene expression signatures in HMA resistant AML cell lines (Leung et al. 2019) , RNA-Seq in patient cohort detected fewer than 0.5% of the total number of differentially expressed genes in non-responders in common for both AZA and DAC. Of note, all the AZA and a large portion of DAC associated genes with down-regulations in non-responders encode immunoglobulins, which is consistent with several recent findings indicating impaired differentiation B cells in association with unfavorable outcomes in MDS and CMML (Ribeiro et al. 2006 and Kahn et al. 2015). We performed flow cytometry analysis in BM cells available for 13 patients prior to start of HMA treatments (5 non-responders and 8 responders), and detected that HMA non-responders had a strong tendency (P=0.07) of decreased baseline frequencies of B cells in their BM than HMA responders.
GSEA analysis based on leading edge genes identified over 200 and 300 biological signaling pathways to be associated with AZA and DAC failure respectively, with 78 pathways commonly correlated with treatment failure of both drugs (28 up-regulated and 60 down-regulated, Table 1). Clustering of these commonly altered pathways based on biological functions revealed that most of them are known to have a role in MDS and CMML pathogenesis and/ or drug resistance. For instance, neurotransmitter, olfactory pathways, and associated G-protein coupled receptor signals was recently reported to play a role in regulating the maintenance and differentiation of BM HSPCs (Shao et al. 2021 and Shim et al. 2013), whereas cell junction signaling that involves integrins and increased MAP2K-MAPK signaling were also reported to be associated with AZA resistance in MDS and CMML (Unnikrishnan et al. 2017).
Among commonly down-regulated biological pathways in non-responders of AZA and DAC, there were clusters of immunoglobulin-associated signals, protein translation regulatory pathways that involve ribosomal proteins, cell cycle signaling, respiratory chain, and Golgi transport signals. Decreased expression of ribosomal proteins and related impairment of ribosomal functions were known mechanism in MDS and CMML development (Ebert et al. 2008 and Schneider et al. 2016). Furthermore, decreased respiratory chain and Golgi transport signals were also identified by transcriptomic investigation in the DAC resistant TF1-RES cell lines.
In addition to the commonly altered signals for both AZA and DAC resistance, innate immune signaling pathways including interferon and toll-like receptor signals were significantly up-regulated in DAC non-responders but down-regulated in AZA non-responders.
CONCLUSIONS: In this study, the transcriptomic data between responders and non-responders to AZA and DAC were investigated separately, thereby drug-specific as well as the common biomarkers associated with treatment failures of both drugs could be identified. The relatively low proportion of genes and pathways shared by AZA and DAC non-responders suggest the difference underlying biological mechanisms of AZA and DAC failure.
Sasaki: Novartis: Consultancy, Research Funding; Pfizer: Membership on an entity's Board of Directors or advisory committees; Daiichi-Sankyo: Membership on an entity's Board of Directors or advisory committees. Wei: Daiichi Sanko: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal